International Journal of Biomedical Science
3(1), 14-19, Mar 15, 2007
© 2005 Master Publishing Group
Overlapping of Genes in the Human Genome
Tomohiro Nakayama1, Satoshi Asai2, Yasuo Takahashi2
Oto Maekawa3, Yasuji Kasama3
1Division of Molecular Diagnostics, Department of Advanced Medical Science, Nihon University School of Medicine, Tokyo, Japan; 2Division of Genomic Epidemiology & Clinical Trials, Department of Advanced Medical Science at the Nihon University School of Medicine, Tokyo, Japan; 3Maize Corporation, Tokyo, Japan
Corresponding Author: Tomohiro nakayama, Division of Molecular Diagnostics, Advanced Medical Resea41rch Center, Ooyaguchi-kamimachi 30-1, Itabashi-ku, Tokyo 173-8610, Japan. Tel: (81-3)3972-8111(ext: 8205);Fax: (81-3)5375-8076; E-mail: tnakayam@med.nihon-u.ac.jp.
|
|
ABSTRACT
 |
Overlapping genes are relatively common in DNA and rNA viruses. there are several examples in bac-terial and eukaryotic genomes, but, in general, overlapping genes are quite rare in organisms other than viruses. there have been a few reports of overlapping genes in mammalian genomes. the present study
identified all of the overlapping loci and overlapping exons in every chromosome of the human genome using
a public database. the total number of overlapping loci on the same and opposite strands was 949 and 743, respectively. similarly, in every chromosome, the instances in which two loci were located on the same strand were similar to the number of 2 genes observed on opposite strands, except for chromosome 5. the number of 2 exons located on the same strand was higher than that for 2 exons located on opposite strands, indicating the presence of many comprehensive-type overlaps. the mean percentage of overlapping exons on opposite strands in each chromosome was 3.3%, suggesting that parts of the nucleotide sequences of 26,501 exons are used to produce 2 transcribed products from each strand. the ratio of the number of overlapping regions to chromosomal length revealed that, on chromosomes 22, 17 and 19, ratios were high for both types of 2 loci, with exons located on the same and opposite strands. ratios were low on chromosomes Y, 13 and 18. these results show that all overlapping types are distributed throughout the human genome, but that distributions differ for each chromosome.
KEY WORDS:
overlapping genes; human genome; locus; exon; chromosome
|
|
INTRODUCTION |
Publication of the human genome sequence marked a
significant milestone in the field of biology. Significant biological
information can also be gained from the analysis
of genome organization. Approximately 30,000 proteincoding
genes are thought to be present in the human genome,
and the positions of genes within chromosomes are
currently being established (1-4).
DNA sequences can code for more than one gene product
by using different reading frames or different initiation
codons. Overlapping genes are relatively common in DNA
and RNA viruses (5-9). While several examples exist in
bacterial and eukaryotic genomes, overlapping genes appear
to be relatively rare in non-viral organisms and few
reports have described overlapping genes in mammalian
genomes (10-12). Some studies have demonstrated that
the overlapping of genes differs among species and have
inferred that this can be attributed to differences in evolutionary
histories (13-15).
Given that both strands of the human genome are used
for transcription, two types of overlapping are thus possible;
2 genes overlapping on the same strand, and 2 genes
overlapping on opposing strands. Furthermore, overlapping patterns can be classified by the relative positions of
the 2 genes. Little exact information is available regarding
overlapping genes in the human genome and their associated
overlapping patterns. To investigate this phenomenon
and the inherent biological information contained therein
further, the number of overlapping genes and their patterns
were examined in every chromosome of the human
genome.
|
|
METHODS
 |
The positions and sequences of each gene were obtained
from the National Center for Biotechnology Information
(NCBI) database (build 31; published January 15,
2003; http://www.ncbi.nlm.nih.gov/genome/guide/human/
HsStats.html). Each locus was defined using both LocusLink
and RefSeq, using gene symbols and names established
by the nomenclature committee for the genome
(http://www.gene.ucl.ac.uk/nomenclature/).
In the LocusLink report, symbols and names were reported
under the banner (http://www.ncbi.nlm.nih.gov/).
Exons were defined as DNA sequences coding mRNA,
rather than considering functions within specific genes or
locations within specific genes. This definition allowed for
analysis of all possible cases of overlapping. Official gene
symbols and names were used as follows: For RefSeq records,
symbols were assigned using the LOCUS system. If
a symbol had not yet officially been assigned, an interim
symbol and name were arbitrarily selected. Arbitrarily
selected symbols and names are included at this website
(http://www.ncbi.nlm.nih.gov/LocusLink/collaborators.
html).
All loci and exons registered in NCBI build 31 were
examined, using the data describing the position of genes
on the chromosome. The information of nucleotide sequences
and the positions of each nucleotide in the whole
human genome were downloaded and stored in EXCEL
file format (Microsoft Corporation, Redmond, Washington).
All overlapping loci and overlapping exons could be
defined according to the start and end positions of each
locus and exon. Data for overlapping loci were produced
using data of registered loci, while the data for overlapping
exons was produced using data distinct coding regions and
mRNA sequences.
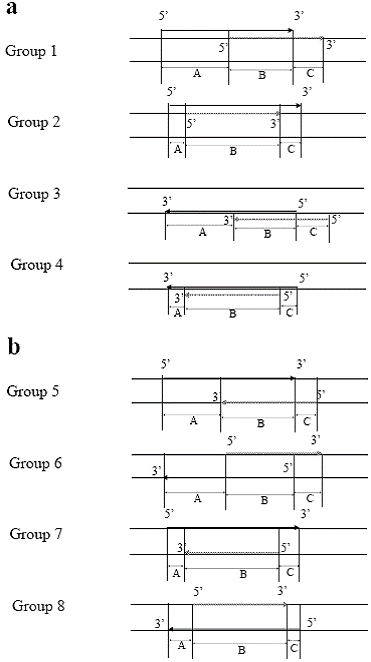
View larger version :
[in a new window] |
Figure 1. Schema of overlapping patterns among loci or exons.
Black arrows indicate locus or exon “1”. Shaded arrows indicate
locus or exon “2”. Locus “2” or exon “2” were defined as those
with the side of short arm (p) were located downstream of locus
1 or exon 1. Figure 1a: Schema of 2 loci or exons on the same
strand. Groups 1 and 2: both regions are on the positive strand
(unidirectional). Groups 3 and 4: both regions are on the negative
strand (unidirectional). Groups 2 and 4: region 1 includes
region 2 (comprehensive). Figure 1b: Schema of 2 loci or exons
on opposite strands. Group 5: convergent; Group 6: divergent;
Groups 7 and 8: comprehensive.
A, the length of the flanking region without overlapping, located
on the side of short arm (p); B, the length of the overlapping
region; C, the length of the flanking region without overlapping,
located on the side of long arm (q).
| |
 RESULTS RESULTS
 |
The type of overlap was classified into 8 groups (Fig.1), with 2 loci or exons on the same strand divided into 4
groups, and the same for those on opposite strands. We classified the patterns of overlapping genes by considering
the strand-location of respective genes. For example, for Groups 1 and 2, both regions are on the positive (sense)
strand and their mRNAs are transcribed using the negative (antisense) strand as the template. Groups 1, 2 and 7
were also different from Groups 3, 4, and 8, respectively.
General information for overlapping loci is shown in
Table 1. A total of 12,692 loci were present on the positive
strand, with 12,442 on the negative strand. The total number of overlapping loci on the same and opposite
strands was 949 and 743, respectively. Except for Group
2 of chromosome 5, the number of instances where 2 loci
were located on the same strand was similar to the number
of 2 loci on opposite strands on every chromosome.
This group on chromosome 5 includes the protocadherin
(PCDH) cluster located on the positive strand of 5q31. The
mean number of overlapping loci was 6.7% of the total
loci on each chromosome (range, 2.0-33.7%). The ratio of
the number of overlapping loci to chromosome length revealed
chromosomal characteristics. On chromosomes 22,
17 and 19, ratios were high when 2 loci were on overlapped
both the same and opposite strands. Analysis of overlap
type revealed Groups 2 and 6 as occurring relatively
frequently.
Given that the organization of several genes has not yet
been clarified, insufficient information is currently available
for determining the incidence/pervasiveness of overlapping
loci in these specific genes. We therefore set about
to examine overlapping exons for the human genome as a
whole.
The total number of exons on the positive and negative
strands was 404,776 and 402,510, respectively (Table
2). The number of 2 exons located on the same strand
(3,843,308) differed substantially from the number of 2
exons on opposite strands (26,501). Interestingly, the number
of 2 exons located on the same strands (groups 2 and 4)
exceeded the number of exons, with comprehensive-type
overlaps (Groups 7 and 8). This indicates the presence
of numerous comprehensive-type overlaps on the same
strands (2 exons located on the same strand with a smaller
exon within the larger exon). The percentage of overlapping
exons (out of the total number of exons) on opposite
strands within each chromosome ranged from 1.1% to
5.5%. The total number of overlapping exons was 26,501
(3.3%) out of 807,286 exons, which suggests that parts of
the nucleotide sequences for 26,501 exons are used to produce
2 transcribed products from each strand.
The ratios for overlapping exons/chromosomal length
on chromosomes 22, 19, 14 and 17 were high for 2 exons
located on both the same or opposite strands. Ratios were
low on chromosomes Y, 13 and 18 for both overlapping
types, suggesting that overlapping is not equally distributed
among chromosomes. The NIT1/DEDD and ARTS-
1/CAST pairs clearly show this overlapping pattern.
 DISCUSSIONS DISCUSSIONS

|
Previous reports (5-12) have counted the number of
genes exhibiting the overlapping phenomenon, but no reports
have enumerated the number of loci or exons that
exhibit this overlap. Furthermore, previous reports (5-12)
have only described this overlap phenomenon for opposite
strands. The present strategy offers a valuable method for
estimating the number of overlapping genes, as the total
number of genes in the human genome is yet uncertain.
Because the total number of genes in the human genome
was estimated 32,000 in 2001 (1, 2), and subsequently estimated
in 2004 to 22,000.
The total number of exons in the human genome has
been estimated at approximately 320,000 (8.8/gene) (1, 2)
whereas the present data indicate the existence of more
than twice that number. This discrepancy is due to different
methods of enumerating exons. We simply counted all
of the exons in the human genome, without considering
how many exons comprise a gene. This method can identify
all exons (e.g., more than 2 exons identified in the same
region) and avoids confusion due to splicing variants.
Overlapping genes may evolve as a result of extensions
of open reading frames (ORF) caused by switching
to an upstream start codon, substitutions in start or stop
codons, or deletions and frame shifts that eliminate initiation
or stop codons (13). The necessity for maintaining
2 functional overlapping genes inevitably constrains the
extent to which both genes can become optimally adapted.
However, such constraints can be alleviated by duplication
of the overlapping gene pair, allowing for independent
evolution of each gene in the resulting copies. This means
that overlapping genes can thus only survive long evolutionary
periods when the overlap confers a selective advantage
upon the organism. In viruses, overlapping genes
are thought to persist due to the considerable constraints
on genome size (7). In non-viral organisms, the potential
advantages of overlapping genes are less clear, although
co-regulation may be involved (4). Results of a comparative
study of overlapping genes in the genomes of two
closely related bacteria revealed that many overlapping
genes arise due to incidental elongation of the coding region
(16). Overlapping genes have generally been thought
to be relatively rare in the human genome, but the results
of the present study show that they are more abundant than
was previously thought. Interestingly, overlapping genes
do not appear to be the result of evolutionary pressure to
minimize the size of the human genome.
Yelin et al. (17) demonstrated by in vitro experiments
that antisense transcription occurs widely in the human
genome. The resulting data set of 2,667 sense-antisense
pairs was evaluated by microarrays containing strandspecific
oligonucleotide probes derived from the region
of overlap. Verification of specific cases by Northern blot
analysis with strand-specific riboprobes confirmed the occurrence
of transcription from both DNA strands. While
these authors also predicted the existence of approximately
1,600 sense-antisense transcriptional units, transcribed
from both DNA strands (13), no overlapping patterns were
elucidated.
Adachi-N et al. (18) reported that some genes overlap
in a head-to-head manner (transcribed in opposite directions),
while Koyanagi-KO et al. (19) recently reported the
occurrence of bidirectional gene pairs in some species.
However, they did not describe the patterns of the overlapping
exons. In our study, this type of overlap was included
in the overlapping loci identified. It has also been reported
that divergence (bidirectionality) is frequently observed,
particularly in genes involved in DNA repair or replication.
(18) The functional significance of this is unclear,
but divergence may permit two genes to share one CpG
island for purposes of coordinated expression. In some
bidirectional loci, expression of two divergent genes has
been found to be coregulated, and promoters exhibiting
bidirectional activity have often been observed (20, 21).
To the best of our knowledge, the phenomenon of overlapping
exons is not specific in DNA repair or replication, and
further studies are needed to clarify the functional significance
of overlapping genes.
Clarification of overlapping genes will facilitate the description
of roles for each strand of the human genome and
will provide insight into the mechanisms of evolution.
These results show that all overlapping types are distributed
throughout the human genome, but that distributions
differ for each chromosome.
|
|
REFERENCES
 |
-
1. Lander ES, Linton LM, Birren B et al. International Human Genome
Sequencing Consortium. Initial sequencing and analysis of the human
genome. Nature 2001;409:860-921.
-
2.Venter JC, Adams MD, Myers EW, et al. The sequence of the human
genome. Science 2001;291:1304-1351.
-
3. Waterston RH, Lander ES, Sulston JE. On the sequencing of the human
genome. Proc Natl Acad Sci USA. 2002;99:3712-3716.
-
4. Istrail S, Sutton GG, Florea L, et al Whole-genome shotgun assembly
and comparison of human genome assemblies. Proc Natl Acad Sci
USA. 2004;101:1916-21.
-
5. Barrell BG, Air GM, Hutchison CA 3rd. Overlapping genes in bacteriophage phiX 174. Nature 1976;264:34-41.
-
6.Noemark S, Bergstrom S, Edlund T, et al. Overlapping genes. Annu
Rev Genet 1983;17:499-525.
-
7. Lamb RA, Horvath CM. Diversity of coding strategies in influenza
viruses. Trends Genet 1991;7:261-266.
-
8. Samuel CE. Polycistronic animal virus mRNAs. Prog Nucleic Acid
Res Mol Biol 1989;37:127-153.
-
9. Keese PK, Gibbs A. Origins of genes: ‘big bang’ or continuous creation?
Proc Natl Acad Sci USA 1992;89:9489-9493.
-
10. Sander C, Schulz GE. Degeneracy of the information contained in
amino acid sequences: evidence from overlaid genes. J Mol Evol
1979;13:245-252.
-
11. Kasper G, Taudien S, Staub E et al. Different structural organization
of the encephalopsin gene in man and mouse. Gene 2002;295:27-32.
-
12. Dan I, Watanabe NM, Kajikawa E, et al. Overlapping MINK and
CHRNE gene loci in the course of mammalian evolution. Nucleic
Acids Res 2002;30:2906-2910.
-
13. Veeramachaneni V, Makalowski W, Galdzicki M, et al. Mammalian
overlapping genes: the comparative perspective. Genome Res
2004;14:280-286.
-
14. McGirr KM, Buehuring GC. Tax & rex: overlapping genes of the Deltaretrovirus
group. Virus Genes. 2006;32:229-239.
-
15.Torresi J. The virological and clinical significance of mutations in the
overlapping envelope and polymerase genes of hepatitis B virus. J Clin
Virol. 2002;25:97-106.
-
16. Fukuda Y, Washio T, Tomita M. Comparative study of overlapping
genes in the genomes of Mycoplasma genitalium and Mycoplasma pneumoniae. Nucleic Acids Res 1999;27:1847-1853.
-
17. Yelin R, Dahary D, Sorek R, et al. Widespread occurrence of antisense
transcription in the human genome. Nat Biotechnol 2003;21:379-386.
-
18. Adachi N, Lieber MR. Bidirectional gene organization: A common
architectural feature of the human genome. Cell 2002;109:807-809.
-
19. Koyanagi KO, Hagiwara M, Itoh T, et al. Comparative genomics of
bidirectional gene pairs and its implications for the evolution of a transcriptional
regulation system . Gene 2 005;353:169-176.
-
20. Platzer M, Rotman G, Bauer D, et al. Ataxia-telangiectasia locus:
sequence analysis of 184 kb of human genomic DNA containing the
entire ATM gene. Genome Res 1997;7:592-605.
-
21. Shimada T, Fujii H, Lin H. A 165-base pair sequence between the
dihydrofolate reductase gene and the divergently transcribed upstream
gene is sufficient for bidirectional transcriptional activity. J Biol Chem
1989;264: 20171-20174.
![]()
![]()